 As described previously, MitoPlex™ is the federation layer of Mitopia’s persistent storage (i.e., distributed database ) model and MitoQuest™ is the principal and most pervasive of the federated containers. MitoQuest™ is directly responsible for persisting data to server collections, stemming and indexing the data, and providing the bulk of Mitopia’s query capabilities. The MitoQuest™ container is built into Mitopia® and pre-registered with MitoPlex™. MitoQuest™ and MitoPlex™ combined are sufficient to give a complete working system for any normal data. It is only necessary to introduce other federation members once multimedia data types are added. The ‘keyCheckFieldType’ plugin registered by MitoQuest™ (see Table 1 – here) lays claim to handling virtually all fields of the Carmot ontology as follows:
As described previously, MitoPlex™ is the federation layer of Mitopia’s persistent storage (i.e., distributed database ) model and MitoQuest™ is the principal and most pervasive of the federated containers. MitoQuest™ is directly responsible for persisting data to server collections, stemming and indexing the data, and providing the bulk of Mitopia’s query capabilities. The MitoQuest™ container is built into Mitopia® and pre-registered with MitoPlex™. MitoQuest™ and MitoPlex™ combined are sufficient to give a complete working system for any normal data. It is only necessary to introduce other federation members once multimedia data types are added. The ‘keyCheckFieldType’ plugin registered by MitoQuest™ (see Table 1 – here) lays claim to handling virtually all fields of the Carmot ontology as follows:
if ( field is a pointer,handle or relative reference to ‘char’)
return YES;
if ( field type is an array of ‘char’ )
return YES;
if ( field is a persistent (‘#’) or collection reference (‘##’ or ‘@@’) )
return YES;
if ( field is descended from integer or real numbers )
return YES;
The logic above means that the MitoQuest™ container will be called upon by MitoPlex™ to handle all field types except memory or relative references to a type other than ‘char’. In other words, multimedia information related code such as images, movies, GIS renderings etc. is all tied into the federation from a field perspective simply through these non-char reference fields. Of course multimedia containers frequently also register additional plugins with MitoPlex™, primarily those involved with server side behaviors. To all intents and purposes however, when we talk about database functionality within Mitopia®, we are talking about MitoQuest™.
Server Folders
Before we can examine how MitoQuest™ operates in a distributed database context, we must first describe the various different folders that can be associated with any given server. These folder types and their purposes derive from Mitopia’s Client/Server abstraction layer and not from the MitoPlex™ layer that is built on that abstraction. It is thus important to describe the various types of server folders in the general case before we get bogged down in the details of their specialized use in a MitoPlex™/MitoQuest™ context. The Client/Server abstraction layer supports the following distinct types of server folders:
The Input Folder
The server input folder is the place where any source and/or multimedia content to be ingested by the server must be placed. Servers with input folders (and not all do), continually monitor the folder (and any sub-folders) for new input. When a new file appears, it is processed and the meta-data associated with the input added to the server’s database. The multimedia content is moved to the server’s output folder for permanent storage. The folder hierarchy seen in the input folder is reflected automatically in the output folder and so by controlling where things appear in the input folder, one can control how the associated multimedia is organized in the output folder. This can be a critical consideration when the server has an associated robotic mass storage device used for archiving data to disk or tape. In MitoPlex™ servers, the input folder may also be used to transfer mined collections of data from clients when those collections are too large to be passed directly through a communication message. In this case the collections are loaded and persisted from the input folder when time is available. This means that it is generally wise for all MitoPlex™ servers to have an associated input folder, even if it is never used for multimedia input and is there simply to handle overly large mined collections.
The Output Folder
The server output folder is the place where the multimedia data associated with the server (if there is any) is stored permanently. Multimedia data (e.g., Image files) arrive in the input folder and is transferred to the output folder as the multimedia file is processed. The organization of the output folder reflects that of the input folder but it contains additional sub-folders which are used to group the multimedia files into a set of numbered ‘batches’. The size of each batch is determined by the nature of any archiving device associated with the server (e.g., a DVD autoloader) and once a batch is complete, if the server is set up to archive, it will be ‘burned’ to archival media and loaded into the autoloader. Once the move to the autoloader is complete, the original archive-related folders in the output folder for the batch are replaced by alias files that reference the media. This means that there is essentially no limit to the amount of data that can be stored in the output folder and that any item that is requested but has been archived is automatically re-loaded from the mass storage system whenever it is requested. The archiving mechanism associated with servers is very powerful and flexible and is not described here. For MitoPlex™ servers (i.e., those associated with the ontology), only a few types generally have associated multimedia files (e.g., images, movies, sounds etc.), and so only those servers require output folders.
The Reject Folder
The server reject folder is the place where multimedia files arriving in the input folder are moved if the server fails to correctly process the file into the output folder. This can happen for a number of reasons, the most common of which is that the source file format changes and the system code or mining script needs to be updated to reflect this change. Source formats frequently change over time, for example the JPEG image format as used by photo-wire sources has changed many times in the past resulting in all incoming photo-wires being moved to the reject folder. After the problem with the new input format has been fixed, all that is necessary is to drag the reject folder contents back into the input folder and allow it to be re-processed. MitoPlex™ servers that do not have associated multimedia files generally do not require reject folders.
The Alias Folder
The server alias folder is frequently associated with those servers that have live feeds. As mentioned previously, server input to be processed must be placed in the server input folder. This can happen in two ways when the source is a live feed. The first way is that the source is some external (i.e., non-Mitopia®) process that produces files as its output and is capable of directly placing those files into the server’s input folder. Most external processes produce output files within their own folder hierarchy and are not capable of placing it in the Mitopia® server input folder. For this reason, Mitopia® provides the ability to follow any alias files that are placed in the alias folder and move any files found in the destination directory into the server’s input file, deleting the originals when it is done. This ability operates over the network which means that even if the live source is running on another machine, its outputs can be automatically moved to the server and ingested. The alias folder can contain multiple aliases and these can be organized in a sub-folder hierarchy which will be reflected into the input folder and which in turn is reflected to the output folder. This allows multiple sources of data to be organized physically by source in the output folder. Servers without live feeds do not require alias folders.
The Collections Folder
The server collections folder is unique to MitoPlex™ (i.e., ontology driven) servers and is the place where all the server data collections and the associated indexing files are stored. The collections folder is organized as a hierarchy which starts from Datum and exactly reflects that portion of the Carmot ontology for which the server is responsible. This means that the folder names and hierarchy of all server collections folders are identical and are driven directly from the ontology. This fact makes it very easy to locate portions of the data held in a server and also to move data manually between servers if necessary. A more complete discussion of the collections folder contents is given later.
The Pending Folder
The server pending folder is associated with the MitoMine™ mining process. When a client machine is performing a mining operation, and that mining script identifies an associated multimedia file (e.g., an image or movie), it moves a copy of that file to the appropriate server’s pending folder within a directory whose name matches that of the mining script. Within the mined collection, the ontological data associated with the multimedia file is tagged to indicate that the file can be found in the pending directory. This behavior is necessary because the mined collection may not be persisted immediately, or it may not be possible when it is persisted for the server to access the original multimedia data on the machine doing the mining. For this reason, the files are cloned into the pending folder so that when the collection is persisted in the server, the corresponding multimedia file can be moved from the pending folder to the input folder and correctly associated with the ontological data to which it relates. In this sense then the pending folder is a temporary repository in the server of multimedia data that will only actually be ingested by the server if and when the corresponding mined ontological collection is persisted by the client machine. Without this mechanism, multimedia files associated with mining runs not performed on the server itself can easily get lost.
The Backup Folder
The server backup folder is used to implement server backup by copying whatever is necessary to backup and restore server state to this folder, tagged by date. Like all server folders, the backup folder itself may be replaced by an alias to a folder on another disk or even across the network. This in conjunction with server backup facility in the “Client/Server Status Window”, allows synchronous system-wide backups and restores for Mitopia-based systems. A complete discussion of this capability will be presented in a later post. All servers should have a backup folder.
MitoQuest™ collections
In the discussion above, we introduced the server collections folder and the fact that it is organized as a folder hierarchy that exactly mimics portion of the Carmot ontology for the system (starting from Datum) for which any given MitoPlex™ server is responsible. This set of collections may in fact be distributed over the drones of a server cluster, and various tools are available for managing this distribution. In this post we will limit ourselves to describing the basics of MitoQuest™ to a level sufficient to allow a full understanding of how the Carmot ontology is leveraged by MitoQuest™ to perform its operations.
Server Collections and Indexes
MitoQuest™ is responsible for creating the server collections folder hierarchy and its contents and for using those contents to implement distributed database functionality. In an earlier post and the associated discussions, we explained how heterogeneous collections of data coming from the client are distributed to the various MitoPlex™ servers involved in the client collection. We also described how each server is responsible only for storing the record types that are equal to or descendant from the key data-type of the server (or master server if clustered) while maintaining references to records that reside on other servers using the unique IDs. Each .COL collection file therefore consists of records of only a single ontological type which corresponds to the folder in which it resides. In order to implement fast search of these collections, MitoQuest™ must also create inverted file index files and rapid lookup recognizers. The various files associated with any given type in the server collections folder hierarchy are summarized in Table 1 below.
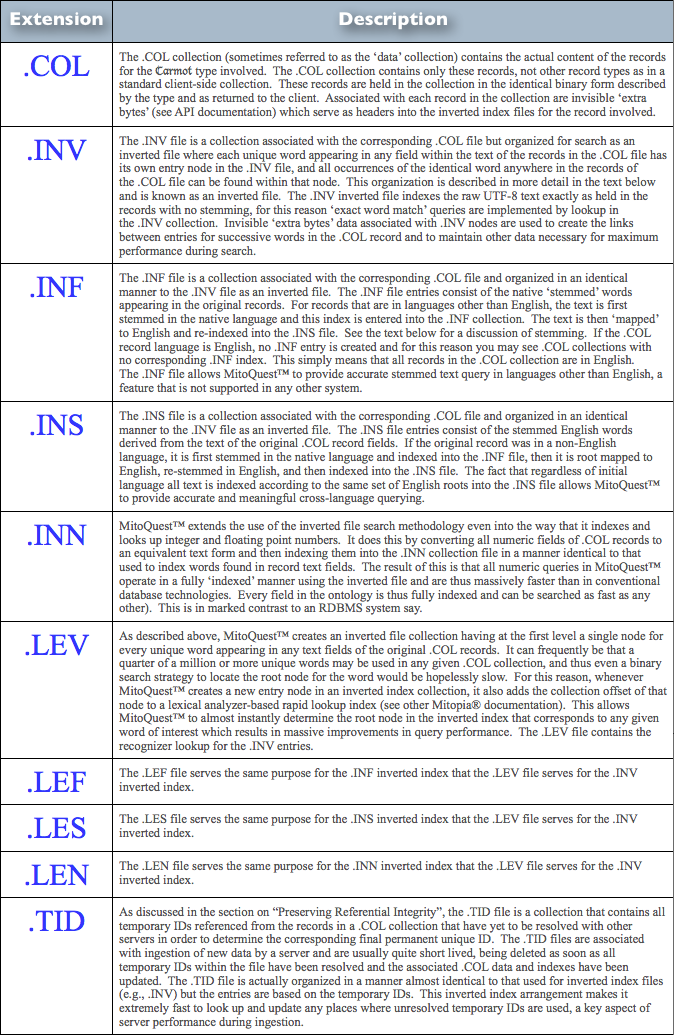 |
| Table 1 – MitoQuest™ Collections Folder File Types (click to enlarge) |
From the descriptions in Table 1, it should be obvious that all the file types other than the .COL files themselves are associated with indexing the information in the .COL file and can thus be re-created at any time provided the .COL file still exists. This process of re-creating the associated files is known as re-indexing and occurs at for example when the ontology is changed or when a backup is restored (backups only contain .COL information). If you manually delete index files and associated lookups, they are created automatically when the application re-starts. In addition, whenever the content of any record in a specific .COL file is altered in any way, the impacted index files (and lookups) must be updated to reflect the changes so that future queries return the correct result. MitoQuest™ handles these operations automatically.
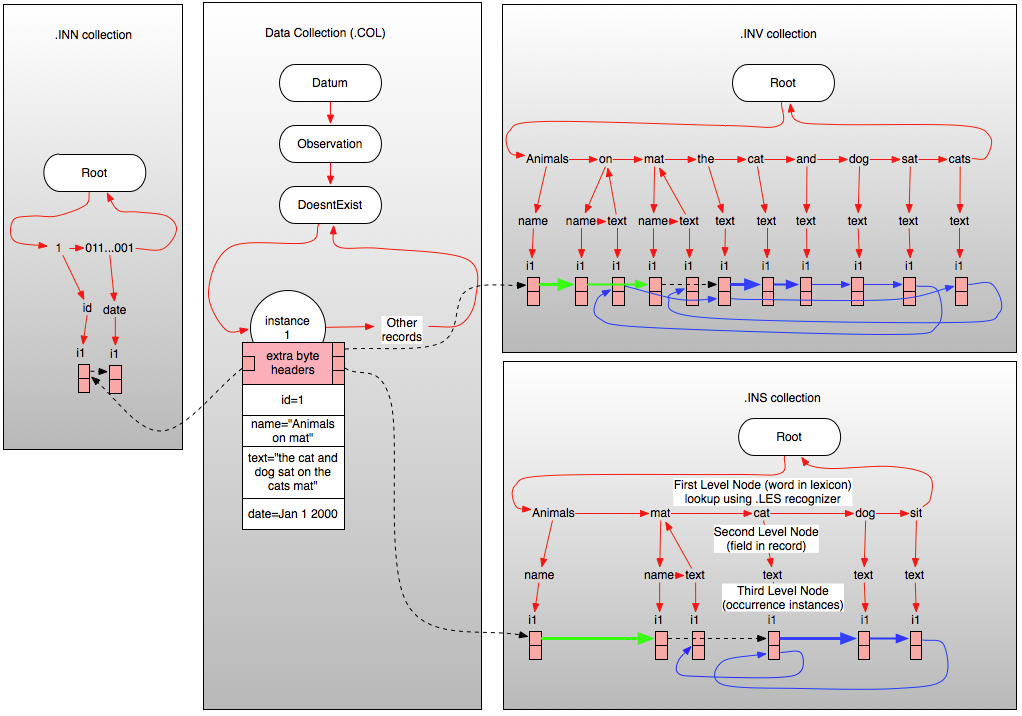 |
| Figure 1 – Relationship Between ‘Data’ collection and inverted index(s) (click to enlarge) |
Figure1 above illustrates a vastly simplified version of the relationships between the server data collection and the .INV, .INS, and .INN index collections in the case of a simple English language record of type “DoesntExist” containing just four fields (which is not possible of course, since all persistent records are derived from Datum) a numeric ID, a date field, a name field (in this case containing “Animals on mat”) and a text field containing the string “the cat and dog sat on the cats mat”.
As you can see from Figure 1, each inverted index file is organized as a three-level tree where the first level node represents (and is named as) the text of the word in the lexicon involved. It is these first level nodes that contain all information regarding a particular word and so all searches essentially start by looking up this node using the lexical recognizer associated with the index and then exploring the nodes within for instances of the word. The second level nodes within each word in the lexicon correspond to (and are named as) the field in Carmot ontological type so that all occurrences of a given word in a specified field can be isolated simply by navigating to the required node at the second level. Finally, the third level nodes contain packed blocks (i1,i2 etc.) each of which contain the details regarding one instance of the word occurring in the specified field. When all the entries in any instance block are used up, a new instance block is created at the same level in the tree (not illustrated). The various inverted file headers associated with each record in the .COL collection point directly to the start of the inverted file entries for that record (the black dotted line), and the inverted file instance records chain through successive fields and within each field through successive words in that field (as illustrated by the green, blue, and dotted black lines shown in the inverted collections). The blue lines in the inverted collections show the word-to-word daisy chain for the ‘text’ field, the green ones show the daisy chain for the ‘name’ field. Black dotted lines indicate field-to-field joining references used by MitoQuest™ to perform “in all fields” queries. In the .INV collection, note that the word “mat” occurs in both the ‘name’ and the ‘text’ fields and so there are two second level nodes below the index node for “mat”. The word ‘and’ in contrast has only one second level node. Also in the .INV collection, note that the word ‘the’ occurs twice in the ‘text’ field and so the daisy chain for the ‘text’ field passes through two entries in the first instance block unlike for all other words.
It should be clear how queries such as “find all records where the text field contains the word dog” can be rapidly implemented. MitoQuest™ simply looks up ‘dog’ in the recognizer, moves to the second level node for the ‘text’ field and then examines all instance records. Even if there were millions of records indexed, if only one record contained the word ‘dog’ then after essentially three reads from the inverted index file, the query engine is able to return the appropriate hit list (by following the links for that entry in the instance slot back to the .COL record offset – not shown). Contrast this to a conventional search that on average would examine 1/2 million records to answer this question. Similarly, it is not hard to imagine how MitoQuest™ might implement sequence queries using this inverted file arrangement for example “find all records where the name field contains the sequence ‘Animals on mat’”; the engine first looks up the entry for “Animals” in the ‘name’ field and then daisy chains through the successive words (shown in green) for the field to compare them with successive words in the query word sequence. In fact, the engine is considerably smarter than this and starts its search at the most statistically uncommon word in the sequence and works forward and backwards (the word-to-word links are actually bidirectional) to match the whole phrase. This trick saves a lot of time in most cases since it reduces the number of records that must be examined, particularly if the query sequence starts with a common word such as “the”.
If we now look at the illustrated connections for the .INS stemmed inverted file we see that it contains considerably fewer nodes at the third level as a result of stemming (see posts on stemming for details). The stemming process has eliminated the words “the”, “on”, and “and” from the ‘text’ field index because they are ‘stop’ words (i.e., should be ignored). The word “cats” has been stemmed to “cat” and thus converted to a second instance of the same word within the ‘text’ field. Finally, the word “sat” has been stemmed to its present-tense form “sit”. If you follow the chain for the ‘text’ field in the .INS inverted file, you will see that for indexing and stemmed query purposes the field contents now reads “cat dog sit cat mat”. Since the same stemming behavior is applied to the query terms issued by the user, it should be clear how this allows MitoQuest™ to support highly accurate stemmed queries, and furthermore how such queries can operate in non-English languages and across languages (by using the .INF and .INS files).
The .INN index in this example contains only two entries, one each for the “id” and “date” fields, but in examples where more numeric fields were involved, this index can be as complex or more complex than the inverted text indices. MitoQuest™ converts numeric fields (real and integer) into the corresponding 64-bit hexadecimal number string, and then creates the .INN inverted file using these strings as the ‘words’ of the lexicon. What this means is that search of all numeric fields both integer and real occurs using an inverted file approach whereby the number is converted to a ‘bucket’ index which gives the top level node in the tree above with which to constrain the search and thus eliminates any time spent examining fields that do not match the criteria thus yielding a massive speed increase. There are of course a number of details involved in this approach that must be solved algorithmically in the search process. Examples are as follows:
- For real numbers, only the integer portion is used to form the bucket index and so if the integer portion exactly matches, it becomes necessary to examine all nodes within the bucket by explicit numeric comparison. For integer numbers, no further numeric test is required since the bucket index exactly represents the value.
- The range of real numbers exceeds that of a 64-bit integer and thus it is necessary to have separate buckets for positive and negative numeric overflow values in addition to the standard integer buckets.
- MitoQuest™ supports signed and unsigned comparisons as well as absolute numeric queries, and this logic adds some complexity to the lookup process.
We have seen how textual and numeric queries are optimized in the MitoQuest™ engine so all that remains is to ask how queries of reference fields (i.e., ‘#’, ‘##’, and ‘@@’ references) are implemented and optimized. For the ‘#’ and ‘##’ fields, the technique used is very similar, that is MitoQuest™ uses the .INN inverted file to index the unique IDs involved in the references. Thus a query like “tell me all people who work for company X or any of its subsidiaries” can be implemented efficiently using just the inverted index files and without requiring explicit examination of consecutive record fields which would otherwise be quite slow. To handle querying fields of ‘@@’ elements (regardless of field type), MitoQuest™ indexes the records into second level field nodes that concatenate the referencing ‘@@’ field path, and the field path within the ‘@@’ element, but ties the third level index records to the ‘@@’ element in the data collection, not the referencing record. This means that in server collections, ‘@@’ elements have ‘extra byte’ entries, and can thus anchor inverted field records. Any hits resulting from ‘@@‘ element indexing are treated as if they belong to the referencing record by QX_AddIfNotPresent(), thereby ensuring correct search results. These inverted index optimizations (and others) mean that MitoQuest™ as a querying engine is capable of significantly outperforming other standard database engines in all areas including ontological level queries (like the “works for” example above) which cannot even be effectively implemented in a taxonomy level database. Combine this with the MitoPlex™ optimizations and federation layer, and the combination can be distributed and scaled to arbitrary levels.
Obviously we have glossed over a huge amount of detail in our discussions of MitoQuest™, however, with the brief descriptions above, it should now be possible to understand how MitoQuest™ persistence and queries tie back to the Carmot ontology for the system. We will discuss this in the next post.

